Research Projects - Axis 2 (PI: L. Etienne)
Axis 2 (PI: Lucie ETIENNE): Functional Evolution of virus-host interactions, and impact on cross-species transmissions
Viruses interact with many cellular proteins, being pro- or anti-viral. Because viruses have been evolving with their host over millions of years, their evolutionary histories include virus-host species coevolution, as well as cross-species transmission events.
Hosts and pathogenic viruses have been locked in an “evolutionary arms-race” or “genetic conflict”. The extent of this conflict is huge, as viruses are major drivers of mammalian adaptation. This notably led to the rapid evolution and signatures of adaptation (positive selection) at the molecular interfaces that have been important in vivo and over time.
Viruses are adapted to their natural host species. However, due to the species-specificity of virus-host interactions, (i) our cellular proteins may be efficient barriers against viral spillovers and (ii) natural variants in other species may be antivirals against human pathogens.
The overall goal of our research program is to discover and to better understand the virus-host molecular interfaces and their evolution. We have a strong focus on viruses that have crossed, or may cross, the species barriers, as well as on mammals that are viral reservoir hosts.
We specifically study evolutionarily- and health-relevant hosts (such as bats, primates, and other mammals) and viruses (such as HIV/AIDS, SARS-CoV, poxvirus…). Although we address our research program in a very fundamental manner, our findings help to inform preventive and therapeutic strategies against current human pathogens and inflammatory genetic diseases, as well as better preparedness against emerging viruses and zoonoses.
Video presenting our lab [in French]: 2024 CNRS Médaille de Bronze
.
Approaches: We use a combination of host and virus phylogenetics and genomics, as well as cellular and molecular biology and virology.
.
Evolutionary & functional characterization of host genes influencing primate lentivirus/HIV
Lentiviruses have coevolved with primates over millions of years, with a mix of coevolution and spillovers, including the cross-species transmissions at the origin of HIV/AIDS in human.
-
We have developed the DGINN pipeline to Detect Genetic INNovations on protein-coding genes (Picard et al. Nucleic Acids Research 2020, publicly available at GitHub). DGINN automates, from a gene name/sequence, the search for homologs in databases, identification of orthologs and paralogs, recombination, and positive selection using five methods. We validated DGINN on 19 primate genes with known evolutionary histories and made some new discoveries along the way! Incollaboration with Laurent Guéguen (LBBE, Lyon), we continue to enrich & update DGINN. -send us your comments!
- We are now using DGINN to characterize the evolutionary history of primate interferon-stimulated genes (ISGs) (Song et al PNAS 2022, Cariou et al PNAS 2022). On the genes with the strongest signatures of adaptation, we perform genetic manipulations and ectopic expression in primate cells to identify the genes influencing the replication of HIV and other primate lentiviruses. Here, we aim to identify and characterize new evolutionarily-relevant HIV-cell interfaces.
.
- We have identified SAMD9L as an interferon-stimulated gene that uses a Schlafen-like box to restrict HIV and inhibit translation in genetic diseases (Legrand et al bioRxiv 2023).
- In collaboration with the Margottin-Goguet Lab, Institut Cochin, we have participated in the discovery and characterization of the HUSH complex and MORC2 as antiviral factors of HIV: Chougui et al Nature Microbiol 2018 , Lasserre et al bioRxiv 2023.
Identifying how ancient and modern viral epidemics have shaped bat genomes, impacting bat immunity and viral emergence
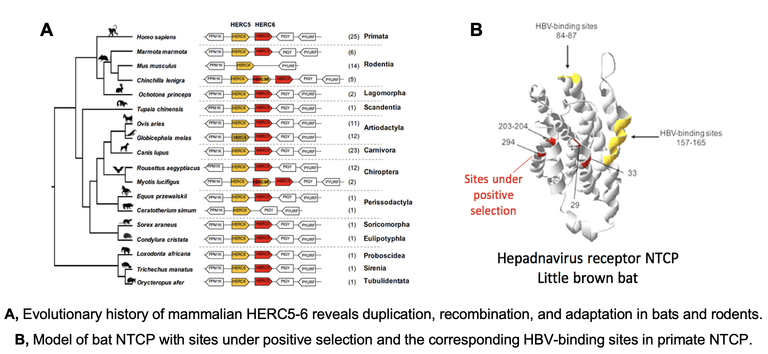
Bats harbor a high number of viruses, but they appear asymptomatic to most viral infections that are pathogenic to other mammals. Amongst other factors, it seems they have evolved a unique balance between immune antiviral defense and viral tolerance. However, most of bat immune evolutionary and functional history remains unknown. Here, we aim to decipher how bat’s immunity has adapted to long-term association with diverse viruses and to determine if and which modern viruses induce a fitness cost on bats. The goals are to determine the impact on modern bat-virus genomes, and leverage natural host genomic variations to identify novel antiviral strategies.
- We uncovered adaptive duplication and functional diversification of Protein kinase R (PKR) that contributed to the uniqueness of bat-virus interactions (Jacquet et al Science Advances 2022 , Prix "Les grandes avancées francaises en biologie" of the French National Academy). See the CNRS press release.
- We characterize the evolutionary and functional diversification of antiviral effectors in bats, in which we found genomic and molecular adaptations. We also hope to bring insights on the genetic determinants of host species-specificity, which partly contribute to current and future epidemics (Jacquet et al Science Advances 2022, Cariou et al PNAS 2022, Jacquet et al Frontiers Immunol 2020, Jacquet et al JVI 2018 ).
.
Funding (in alphabetical order):
amfAR, ANR, ANRS, CNRS, FINOVI, FRM, IRC CNRS-U.Arizona, LabEx ECOFECT, Sidaction
Alumni:
2022-2023 - Clara Loyer, Assistant Engineer.
2016-2023 - Stéphanie Jacquet, ANR/Labex Postdoc. Comentored with D. Pontier. Now, CNRS faculty @ MIVEGEC Montpellier.
2022 - Clément de La Myre Mory, Assistant Engineer. Now, Research tech @ Bruxelles, Belgium.
2021-2022 - Clara Dahoui, Assistant Engineer.
2016-2020 - Léa Picard, PhD student, ENS-Lyon. Comentored with L. Guéguen. Now, Postdoc @ Tadashi Yamamoto Lab, Okinawa, Japan.
2020 - Clément Verez, Undergraduate student, IUT, Université Lyon 1. Now, Student @ ENS-Lyon.
2019 - Margaux Pillon, Master-degree student, M2 "Infectiologie", UCBL. Now, PhD student @ UCBL.
2019 - Manon Picquenot, Master-degree student, M2 "Biodiversité, Ecologie et Evolution", UCBL.
2015-2019 - Mégane Wcislo, Graduate student, UCBL and ENS Lyon.
2016-18 - Fabien Filleton, Ingénieur de Recherche. Now, Engineer.
2017-18 - Ariel De Bernardo, Assistant Ingénieur. After, Ingénieur d'Etude @ CRCL Lyon.
2016 - Nina Courault, Master-degree student, University of Tours.
2014 - From the Emerman Lab: Rossana Colon-Thillet, Prep Student, University of Washington. After, PhD student in the Malik Lab @ Fred Hutch, Seattle WA, USA.