What do we do?
Around 250 million individuals are chronically infected by hepatitis B virus (HBV) and at least 20 million of them are chronically infected with both HBV and hepatitis Delta viruses (HDV). This co-infection is one of the most prevalent worldwide and lead to the most aggressive chronic form of viral hepatitis, with an accelerated progression towards fibrosis/cirrhosis and an increased risk of liver failure, liver cancer and death. Current clinically accepted antiviral treatments for HBV generally lead to a transient or long-lasting reduction of viremia in the blood of patients but without clearance of the virus. Moreover, management of chronic hepatitis delta remains mostly empiric since there are currently no specific treatments and that the use of Pegylated interferon-alpha (Peg-IFN-α) is inefficient for most patients.
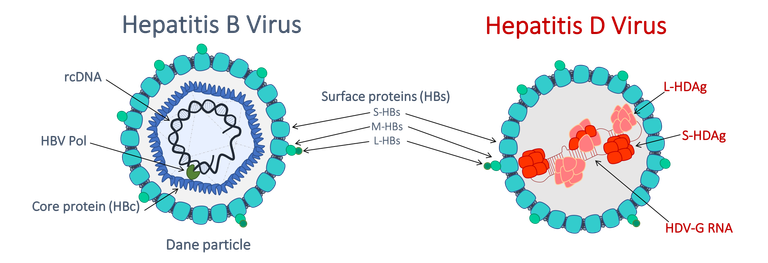
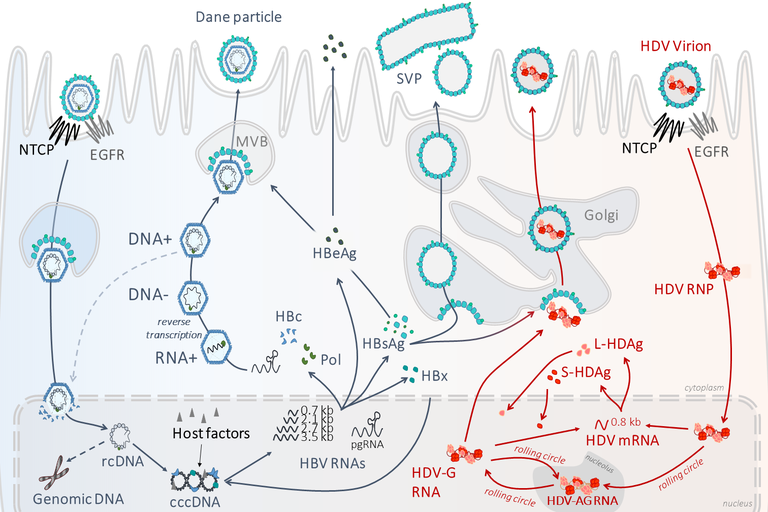
HBV functionally persists in the nucleus of hepatocytes as a non-integrated, episomal DNA, the so-called cccDNA. The HBV cccDNA serves as template for transcription of all HBV RNAs by the cellular RNA polymerase II (RNA Pol-II) thereby driving the production of viral antigens such as HBeAg, HBsAg and progeny virions (containing a partially double stranded relaxed circular DNA that results from reverse transcription of a pregenomic RNA within assembled nucleocapsid). The HBV cccDNA molecule is organized into a chromatin-like structure that displays the typical beads-on-a-string arrangement in electron microscopy (Bock, Schranz et al. 1994, Newbold, Xin et al. 1995). Viral proteins (HBx, HBc), host histones but also non-histone cellular proteins are directly or indirectly attached to cccDNA (Bock, Schwinn et al. 2001, Belloni, Pollicino et al. 2009, Lucifora, Arzberger et al. 2011) and its transcriptional activity is subjected to the “histone code” (Pollicino, Belloni et al. 2006, Belloni, Pollicino et al. 2009, Benhenda, Ducroux et al. 2013).
HDV virions contain a ribonucleoprotein composed of a circular single-stranded negative RNA genome, presenting a “quasi” double-stranded conformation, and of viral proteins called HDAg. HDV is a defective satellite virus that uses the envelope of HBV (Rizzetto, Canese et al. 1980) to egress from and to re-enter into hepatocytes. Apart from those steps, HDV intracellular RNA replication is thought to be independent of HBV. In the nucleus of infected cells incoming genomes serve as template for the synthesis of replicative intermediates called anti-genomes (AGs), which are fully complementary to HDV genome sequence and bear the ORF for HDAg proteins expression. AGs originate from genomic template by the action of DNA-dependent/RNA-polymerase II (RNA Pol-II) and/or Pol-I, and vice versa (AG→G), new genomes are generated from AGs (AG→G), via a a rolling circle process involving viral ribozyme activities. HDV mRNAs are transcribed from Gs (Hsieh, Chao et al. 1990, Gudima, Wu et al. 2000) and translated into the HDV proteins (S- and L-HDAg).
--------
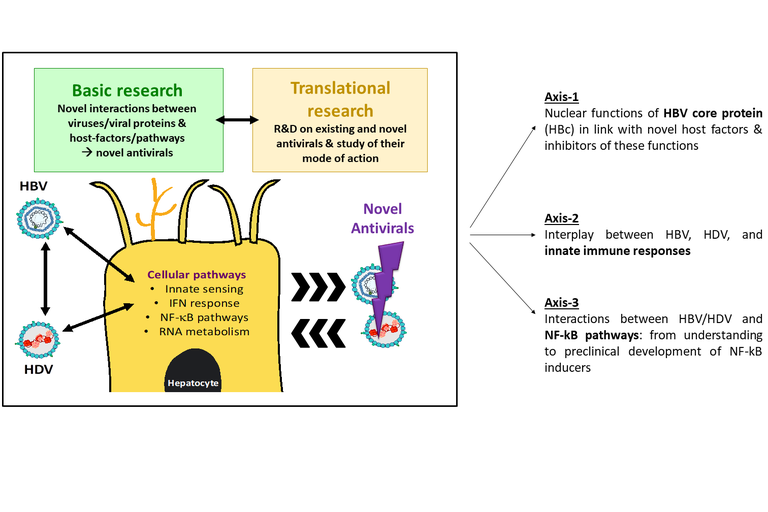
The objectives of our team are to further gain original and relevant knowledge on the interactions between HBV/HDV viruses with host-factors in liver cells (i.e., hepatocytes, liver resident innate immune cells) in order to better (1) understand the early steps leading to viral establishment and persistence, (2) characterize viro-induced mechanisms of liver pathogenesis using relevant and newly-developed animal models, (3) identify novel targets/biomarkers for the development of novel host-targeting antiviral agents (HTA), which could also be interesting to prevent pathogenesis progression and onset/progression of hepatocellular carcinoma. Our research program, that include interconnected basic and translational research, is divided into three main axes as shown in the figure below.
Our research is supported by the ANRS